The Australian Tree of Life Bioinformatics team (AToL Bioinformatics) is developing infrastructure for the rapid generation and publication of genome assemblies and annotations. The current focus of AToL Bioinformatics is optimising and automating these processes in the Genome Engine.
Our Genome Engine was inspired by the Wellcome Sanger Institute’s Genome Engine, and uses some of their pipelines under the hood.
You can learn more about the Australian Tree of Life activity here.
What does the Genome Engine do?
The Genome Engine is a semi-automated workflow for assembling and annotating genome sequences from raw sequence data, brokering data to International Nucleotide Sequence Database Collaboration (INSDC) repositories, and drafting short Genome Notes.
This involves:
- Ingesting raw sequence data from the Bioplatforms Australia Data Portal
- Processing sampling and sequencing metadata
- Assembling genome sequences from sequence read data
- Annotating assembled genomes
- Brokering sample metadata, sequence reads, and genome assemblies to the European Nucleotide Archive (ENA)
- Generating an automatic Genome Note providing details and metrics about sampling, sequencing and assembly
At present, the Genome Engine is configured to ingest data generated as part of Bioplatforms Australia’s Framework Initiatives, which are available from the Bioplatforms Australia Data Portal. In future, we intend to make the Genome Engine available to any Australian researcher for use with their own sequencing data.
How does the Genome Engine work?
Data retrieval and processing
The Genome Engine accesses sequence data and metadata in bulk from the Bioplatforms Australia Data Portal API. The metadata are provided by the collecting researcher and sample preparation and sequencing facilities.
Packages are filtered to select those relevant to genome assembly and annotation, and metadata are validated and mapped to an intermediary, INSDC-compliant schema.
Taxon and sample identifiers are extracted to determine which packages can be combined in the assembly process and to retrieve species information from the NCBI Taxonomy.
Genome assembly and annotation
Sequence read data are processed and assembled on High-Performance Computing (HPC) facilities at the Pawsey Supercomputing Research Centre, provided by the Australian BioCommons Leadership Share (ABLeS) program.
The assembly pipeline used is an adaptation of the Sanger Tree of Life (ToL) assembly pipeline, which includes the following steps:
- assembly using hifiasm
- redundant contig removal with purge_dups
- optional haplotype resolution with hifiasm and scaffolding with YaHS if Hi-C data is available
Oxford Nanopore-based contig building and scaffolding are currently in development, along with quality assessment and annotation of assembled genomes.
Data brokering
The data broker component of the Genome Engine uses sample, sequencing, and assembly metadata to submit files automatically to the ENA. BioSample information is submitted using the ToL sample checklist, a minimum standard for sample metadata devised by the Darwin Tree of Life project to facilitate data contextualisation and interoperability. Experiment, read, and assembly data are submitted according to ENA’s standards and schemas. In order to comply with these standards, certain metadata fields in the original Bioplatforms metadata must be filled and vocabulary terms used (see the FAQ for more information about metadata requirements). The Genome Engine’s metadata mapping processes allow for these metadata to be formatted in XML files for programmatic submission to the ENA.
The submitted XML files include the data release date, which is determined according to the embargo release date specified in the Bioplatforms data portal. Once records are made public on their release date, they are exchanged with and made available from other INSDC databases at the US National Center for Biotechnology Information (NCBI) and the DNA Data Bank of Japan (DDBJ).
Genome Note generation
Once a genome has been assembled, a Genome Note document is generated, outlining key metadata and assembly metrics. The Genome Note pipeline populates a template document with metadata values relating to taxonomy, specimen collection, nucleic acid extraction, sequencing, and assembly, and key metrics calculated in the assembly pipeline. The Genome Note also contains the accession numbers generated during brokering to the ENA. The project lead and project collaborators (as they are listed in the Bioplatforms metadata) are named as first and second authors.
Genome Notes will be made available to researchers prior to release to provide an opportunity to manually edit and add content.
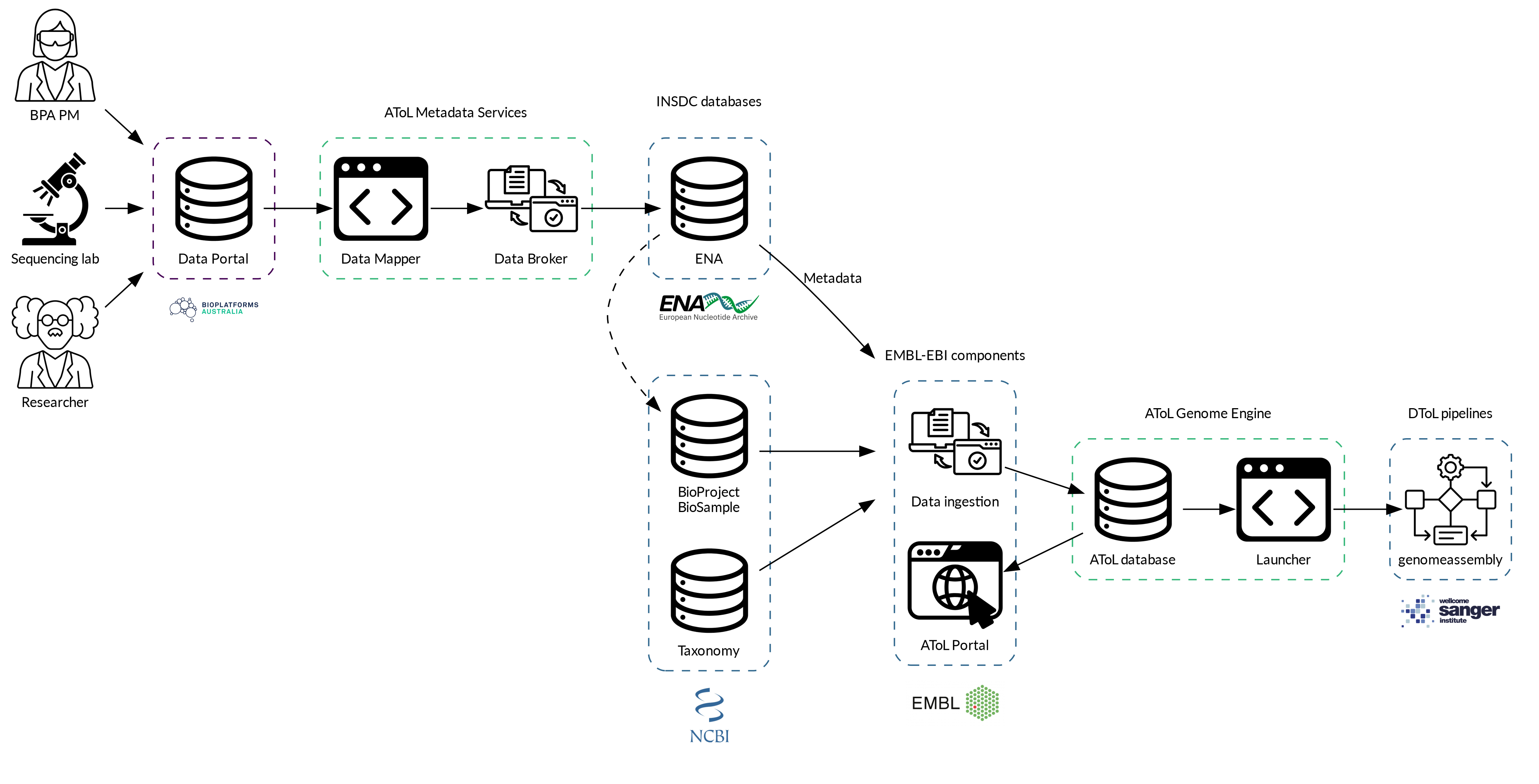 Genome
Engine data flow.
Genome
Engine data flow.
Partners
AToL Bioinformatics is co-funded by Bioplatforms Australia and the Minderoo Foundation, with project partners at the University of Melbourne, QCIF, the European Nucleotide Archive, and the Darwin Tree of Life. The AGRF are hosting a PhD student intern.
Bioplatforms Australia is enabled by NCRIS.
Funding partners
Project partners
PhD industry placement partner
Acknowledgements
This documentation page makes use of the ELIXIR toolkit theme.








