If you are new to the ABLeS program, please read the processes below to get started using ABLeS resources. You may also contact us for more information and assistance.
Which ABLeS allocation matches your work?
Allocation Creation of reference data assets
ABLeS reference data allocations support research groups and consortia within the life sciences to access the dedicated compute capacity required to efficiently construct reference data.
Allocation Production bioinformatics
Institutes, consortia and core facilities are increasingly facing issues scaling their in-house compute and data infrastructure to the questions, sample sizes, and data set sizes they are addressing as part of their research programs. ABLeS production allocations support these groups to implement and run their computational workflow approaches for omics data analysis at scale.
Allocation Software accelerator
Software accelerator allocations will directly support the further development, installation, optimisation, testing and/or benchmarking of bioinformatics software. These allocations are intended to create a culture of best practice in software, helping bioinformaticians to effectively share and document their work, and make it FAIR (findable, accessible, interoperable and reusable).
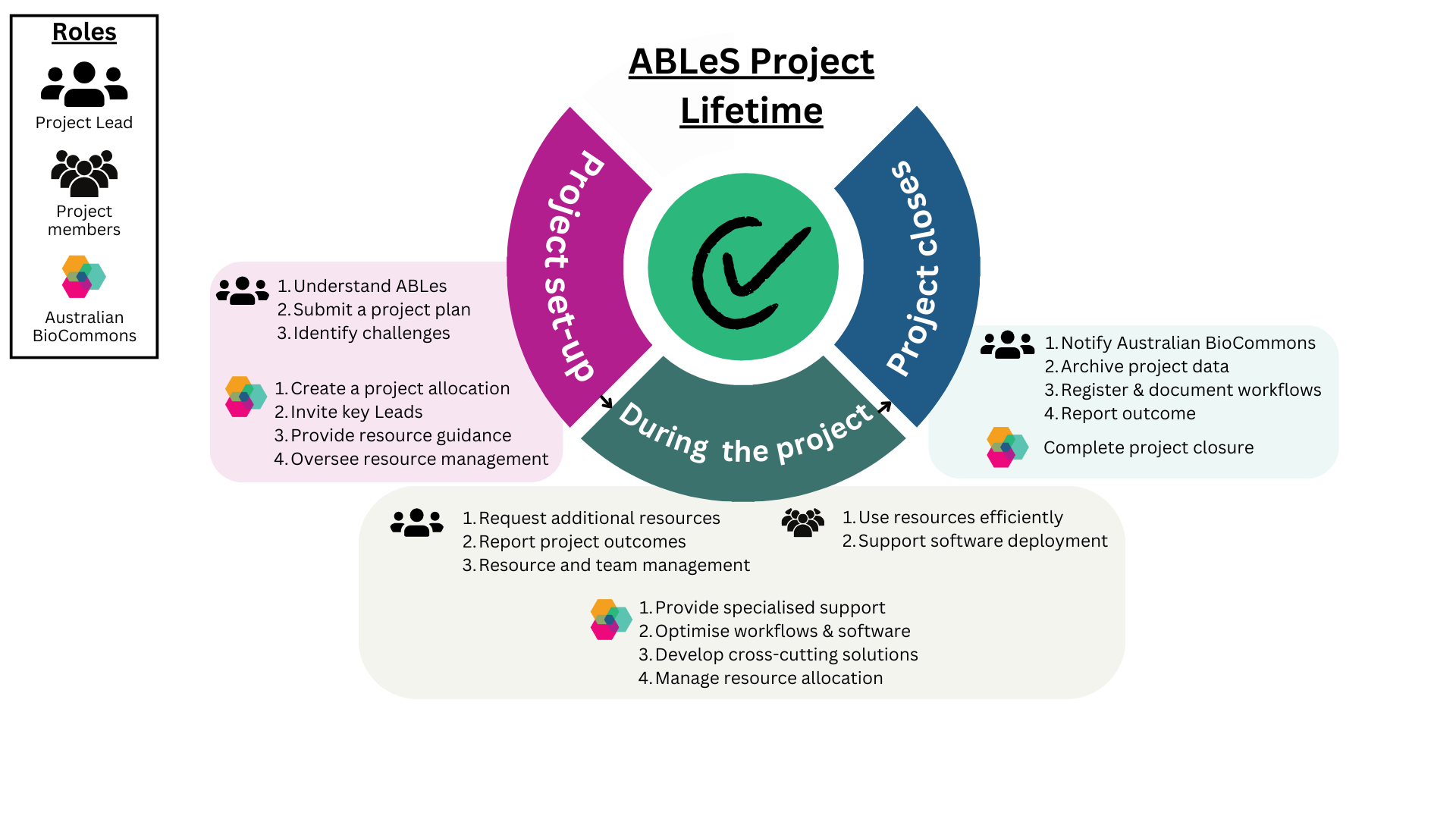
ABLeS process for different stages of a project’s lifetime
Australian BioCommons and the bioinformatics leads for each group have different roles during an ABLeS project’s initiation, operation and closing phases.
ABLeS projects are led and maintained by their users with the support of the ABLeS team to facilitate access to ABLeS resources.
Use the drop-down menu below to explore more details:
- Familarise themselves with ABLeS schemes, expectations, and responsibilities.
- Submit a project plan that contains the details of the project (link). The community’s steering committee (or bioinformatics leads) should approve the plan.
- Identify known challenges that BioCommons, NCI, or Pawsey may be able to address and / or support as part of ABLeS. You can use the GoogleForm to let us know about these challenges.
- Create of a project allocation (at NCI or Pawsey) for a group
- Invite the group bioinformatics leads to a project allocation as Chief Investigators
- Provide information on how to utilise ABLeS resources and contribute to the shared tool and software repository.
- Manage the resources available to the ABLeS program across all active projects.
- Request additional resources when the project needs more resources than are available. Each project gets default resources automatically at the beginning of each quarter without the need to request them.
- Attend a quarterly meeting with BioCommons to discuss and report the outcomes of the project (data, methods, publications etc.) in the previous quarter.
- Manage the resources provided by ABLeS including:
- Adding members to the project(s).
- Educating / onboarding new project members.
- Contributing and / or coordinating contributions to the shared tool and software repository (if89), as well as encouraging community contribution.
- Utilise ABLeS resources efficiently and use the resource for the project purposes.
- Help to deploy and install software, tools and workflows to the shared repository (this activity may be coordinated by the project bioinformatics leads, as needed)
- Installing tools, software, pipelines when special support is needed.
- Optimising workflows and software to be used by all projects.
- Developing and improving cross-cutting tools and workflows.
- Managing the allocation of resources for projects and adding default resources at the beginning of each quarter.
- Notify Australian BioCommons when you are ready to close your project.
- Make sure all project data is archived as the project storage will be released.
- Make sure all workflows are registered with WorkflowHub and well documented so they are findable.
- Report the project outcomes to Australian BioCommons.
- Finalise the closing process with the infrastrucure provider.