Description
HiFi-assembly-workflow is a bioinformatics pipeline that can be used to analyse Pacbio CCS reads for de novo genome assembly using PacBio Circular Consensus Sequencing (CCS) reads. This workflow is implemented in Nextflow and has 3 major sections.
Please refer to this documentation for detailed recommendations relevant to each workflow section:
How-to cite this workflow
citation goes here
Diagram
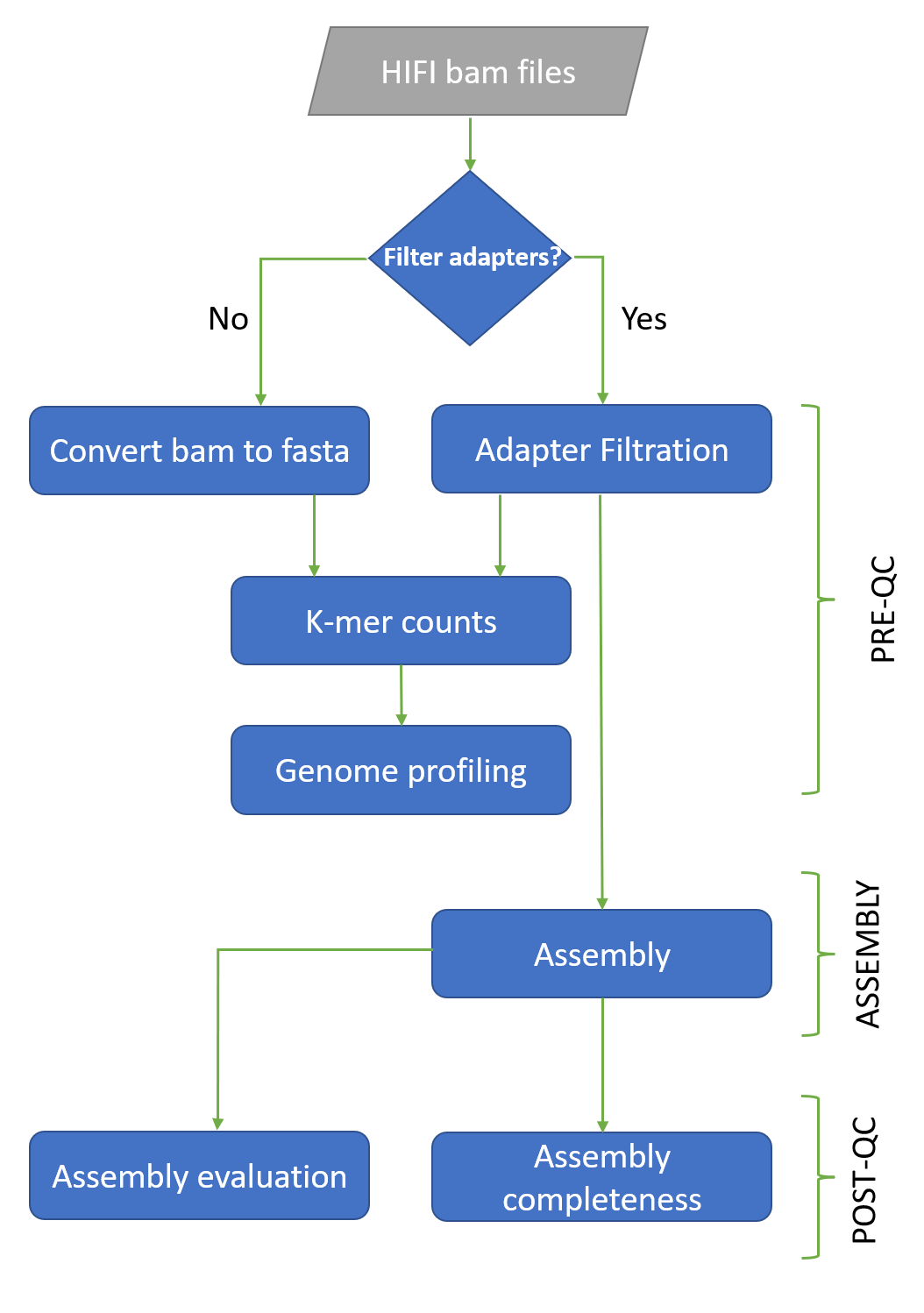
User guide
Quick start guide
The pipeline has been tested on NCI Gadi, Setonix Pawsey, AWS and AGRF balder cluster. If needed to run on AGRF cluster, please contact us at bioinformatics@agrf.org.au.
Please note:
- For running this on NCI Gadi you need access. Please refer to Gadi guidelines for account creation and usage.
- For running this on Setonix Pawsey you need access. Please refer to Setonix guidelines.
- That you can either run jobs interactively or submit jobs to the cluster. This is determined by the
-profileflag. By passing theif89/setonixtag to the profile argument, the jobs are submitted and run on the cluster and also use singularity containers.
For support accessing compute infrastructure, please refer to ABLeS documentation at https://australianbiocommons.github.io/ables/.
Required (minimum) inputs/parameters
PATH to HiFi bam folder is the minimum requirement for the workflow.
Parameters
The workflow accepts the following arguments:
Mandatory arguments
--bam_folder: Folder containing BAM files (Only HiFi BAM file)
Optional arguments
--out_dir: Path to the otuput directory. Default: The input bam directory.--samtools_threads: Number of threads to use for samtools. Default is 8.--samtools_memory: Memory to use for samtools. Default is 16.G.--adapter_filtration: Apply adapter Filtration on the bam files. Default is false.--hifi_adapter_threads: Number of threads used by hifi_adapter. Default is 16.--hifi_adapter_memory: The memory to use for hifi_adapter. Default is 24.--jellyfish_mer_len: The mer length for jellyfish. Default is 21.--jellyfish_threads: The number of threads used by jellyfish. Default is 20.--jellyfish_hash_size: The hash size used by jellyfish. Default is 1G.--jellyfish_memory: The memory used by jellyfish. Default is 24.G.--genomescope_threads: The number of threads used by genome-cope. Default is 1.--genomescope_memory: The memory used by genomescope. Default is 24.G.--ipa_jobs: Number of jobs used by ipa. Default is 4.--ipa_threads: Number of CPU to run ipa. Default is 8. The number of the requested cpus willipa_jobs * ipa_threads.--ipa_memory: The memory used by ipa. Default is 32.G.--quast_threads: The number of threads used by quast. Default is 32.--quast_memory: The memory used by quast. Default is 24.G.--busco_lineage_dir: The path to busco database. if not provided or set to [] busco will download the lineage. default on if89 profile is Default onif89is/g/data/if89/data_library/busco_db/14082023/.--busco_lineage: The lineage to use from the busco database. default iseukaryota.--busco_threads: The number of threads used by busco. Default is 32.--busco_memory: The memory requested for busco. Default is ${params.busco_memory}.--project_id: Project id on GADI to submit jobs through. Mandatory when using if89 profile. (Example: xl04 for AusARG).--storage_paths: Storage_paths on GADI to to be accessed from the compute nodes. Mandatory when using if89 profile. (Example: gdata/if89+gdata/xl04).-
--singularity_cache: The path to the singularity local cache directory. Default onif89is/g/data/if89/singularity_cache/ --aws_execution_role: The execution role to be used when running the pipline using aws batch on AWS cloud service. Mandatory when usingawsprofile.--aws_region: The AWS region needed for configuring running the pipline using aws batch on AWS cloud service. Mandatory when usingawsprofile.--aws_queue: The AWS batch queue to submit the jobs to. Mandatory when usingawsprofile.
Nextflow arguments:
-profile: execution profiles.if89/balder/local/setonix/aws/singularity/docker/
Third party tools / dependencies
The following packages are used by the pipeline.
nextflow/21.04.3samtools/1.12jellyfish/2.3.0genomescope/2.0ipa/1.3.1quast/5.0.2busco/5.4.3HiFiAdapterFilt/2.0.0
The following paths contain all modules required for the pipeline.
/apps/Modules/modulefiles/g/data/if89/apps/modulefiles/
Infrastructure usage and recommendations
Please see the infrastructure documentation.
Outputs
Pipeline generates various files and folders here is a brief description:
The pipeline creates a folder called secondary_analysis that contains two sub folders named:
exeReportResults– Contains preQC, assembly and postQC analysis files
exeReport
This folder contains a computation resource usage summary in various charts and a text file.
report.html provides a comprehensive summary.
Results
The Results folder contains three sub-directories preQC, assembly and postqc. As the name suggests, outputs from the respective workflow sections are placed in each of these folders.
preQC
The following table contains list of files and folder from preQC results
| Output folder/file | File | Description |
|---|---|---|
| Bam files converted to fasta format | ||
| kmer_analysis | Folder containing kmer analysis outputs | |
| k-mer counts from each sample | ||
| histogram of k-mer occurrence | ||
| genome_profiling | genomescope profiling outputs | |
| summary.txt | Summary metrics of genome scope outputs | |
| linear_plot.png | Plot showing no. of times a k-mer observed by no. of k-mers with that coverage |
Assembly
This folder contains final assembly results in
<sample>_primary.fa- Fasta file containing primary contigs<sample>_associate.fa- Fasta file containing associated contigs
postqc
The postqc folder contains two sub folders
assembly_completenessassembly_evaluation
assembly_completeness
This contains BUSCO evaluation results for primary and associate contig.
assembly_evaluation
Assembly evaluation folder contains various file formats, here is a brief description for each of the outputs.
| File | Description |
|---|---|
| report.txt | Assessment summary in plain text format |
| report.tsv | Tab-separated version of the summary, suitable for spreadsheets (Google Docs, Excel, etc) |
| report.tex | LaTeX version of the summary |
| icarus.html | Icarus main menu with links to interactive viewers |
| report.html | HTML version of the report with interactive plots inside |
Compute resource usage across tested infrastructures
NCI Gadi
Computational resource for plant case study
| Time | CPU | Memory | I/O | ||||
|---|---|---|---|---|---|---|---|
| Process | realtime | realtime | %cpu | peak_rss | peak_vmem | rchar | wchar |
| Converting bam to fasta for sample | 12m 48s | 12m 48s | 99.80% | 5.2 MB | 197.7 MB | 43.3 GB | 50.1 GB |
| Generating k-mer counts and histogram | 26m 36s | 26m 36s | 1725.30% | 19.5 GB | 21 GB | 77.2 GB | 27.1 GB |
| Profiling genome characteristics | 13.2s | 13.2s | 89.00% | 135 MB | 601.2 MB | 8.5 MB | 845.9 KB |
| Denovo assembly | 6h 51m 11s | 6h 51m 11s | 4744.40% | 84.7 GB | 225.6 GB | 1.4 TB | 456 GB |
| evaluate_assemblies | 4m 54s | 4m 54s | 98.20% | 1.6 GB | 1.9 GB | 13.6 GB | 2.8 GB |
| assemblies_completeness | 25m 53s | 25m 53s | 2624.20% | 22 GB | 25.2 GB | 624.9 GB | 2.9 GB |
Computational resource for bird case study
| Time | CPU | Memory | I/O | ||||
|---|---|---|---|---|---|---|---|
| Process | realtime | realtime | %cpu | peak_rss | peak_vmem | rchar | wchar |
| Converting bam to fasta for sample | 7m 9s | 7m 9s | 86.40% | 5.2 MB | 197.8 MB | 21.5 GB | 27.4 GB |
| Generating k-mer counts and histogram | 15m 34s | 15m 34s | 1687.70% | 10.1 GB | 11.7 GB | 44 GB | 16.6 GB |
| Profiling genome characteristics | 1m 15s | 1m 15s | 15.30% | 181.7 MB | 562.2 MB | 8.5 MB | 819.1 KB |
| De novo assembly | 9h 2m 47s | 9h 2m 47s | 1853.50% | 67.3 GB | 98.4 GB | 1 TB | 395.6 GB |
| evaluate assemblies | 2m 48s | 2m 48s | 97.50% | 1.1 GB | 1.4 GB | 8.7 GB | 1.8 GB |
| assemblies completeness | 22m 36s | 22m 36s | 2144.00% | 22.2 GB | 25 GB | 389.7 GB | 1.4 GB |
Benchmarking
N/A
Additional notes
N/A
Help/FAQ/Troubleshooting
Direct training and help is available if you are new to HPC and/or new to NCI/Gadi.
- Basic information to get started with the NCI Gadi for bioinformatics can be found here.
- For NCI support, contact the NCI helpdesk.
- Queue limits and structure are explained here.
3rd party Tutorials
- A tutorial by Andrew Severin on running GenomeScope 1.0 is available here.
- Improved Phased Assembler tutorial is available here.
- Busco tutorial.
Licence(s)
MIT License
Copyright (c) 2022 AusARG
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
Acknowledgements/citations/credits
Jung, H. et al. Twelve quick steps for genome assembly and annotation in the classroom. PLoS Comput. Biol. 16, 1–25 (2020).
2020, G. A. W. No Title. https://ucdavis-bioinformatics-training.github.io/2020-Genome_Assembly_Workshop/kmers/kmers.
Sović, I. et al. Improved Phased Assembly using HiFi Data. (2020).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Waterhouse, R. M. et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548 (2018).