The European Genome-phenome Archive (EGA) (Freeberg et al., 2022) is a controlled access data repository for sensitive human data. The repository is managed by EMBL-EBI (UK) and the Centre for Genomic Regulation (CRG) (Spain). Submission is open to anyone and data access is controlled per dataset by a user submitted data access policy and user defined data access committee. A single data access committee and policy may be used for a single dataset or may be reused for multiple datasets.
EGA Submission process
The EGA submission process can be challenging to those unfamiliar to it, so the following sections provide an overview as well as some tips and tricks that may help you along the way. The first step is to gain an EGA submission account, which can be done by completing the submission account form.
EGA Metadata
The metadata files that must be submitted to EGA follow the same INSDC schemas, with the addition of policy, dataset and dac objects. These allow for the administration aspects related to controlled access of data and allow for finer grained access to datasets within a study. All information submitted in these XML files is considered public access and can be queried via the EGA API once the study is available in the platform. The required attributes that must be included about each sample are:
- Donor ID - anonymised individual identifier
- Biological sex (referred to as gender)
- Phenotype - preferably ontologised using Experimental Factor Ontology (EFO)
Submitters can add as many other attributes as they like, but this information will be openly accessible when the study is submitted. It is also possible to first submit attributes to the BioSamples database first and use the obtained accessions in your EGA submission to link to BioSamples. These would also only be for public metadata.
Due to limited required attributes, it is often necessary to make more comprehensive clinical, phenotypic and demographic metadata available alongside the data files. If this metadata is subject to controlled access, the information can be attached to the submission as Analysis (phenotype) files. These do not follow any kind of strict standard or format, however the use of Phenopackets is an emerging standard for characterising this type of information. README files can also be incorporated to provide an explanation of the phenotype or other analysis files.
Submitting to the EGA can be done via its submission interface or through submitting XMLs or JSONs via their REST API. The submission interface is useful for a user-friendly experience when submitting metadata for objects where there are relatively few objects of that type. We recommend using it for objects such as:
- Study
- DAC
- Policy
- Experiment
For objects with a high cardinality, such as Samples, Runs and Analyses, using the CSV templates that are available through the submission portal or creating XML documents and submitting via API is more efficient.
However, it is possible to create a fully automated submission process that contains all metadata required by creating XML documents.
Submission notes:
Table: Detailed information about ega objects and tips on creating XMLs for each object. Adapted from (Band, 2019; EGA, 2022)
Table to be reformatted and placed here.
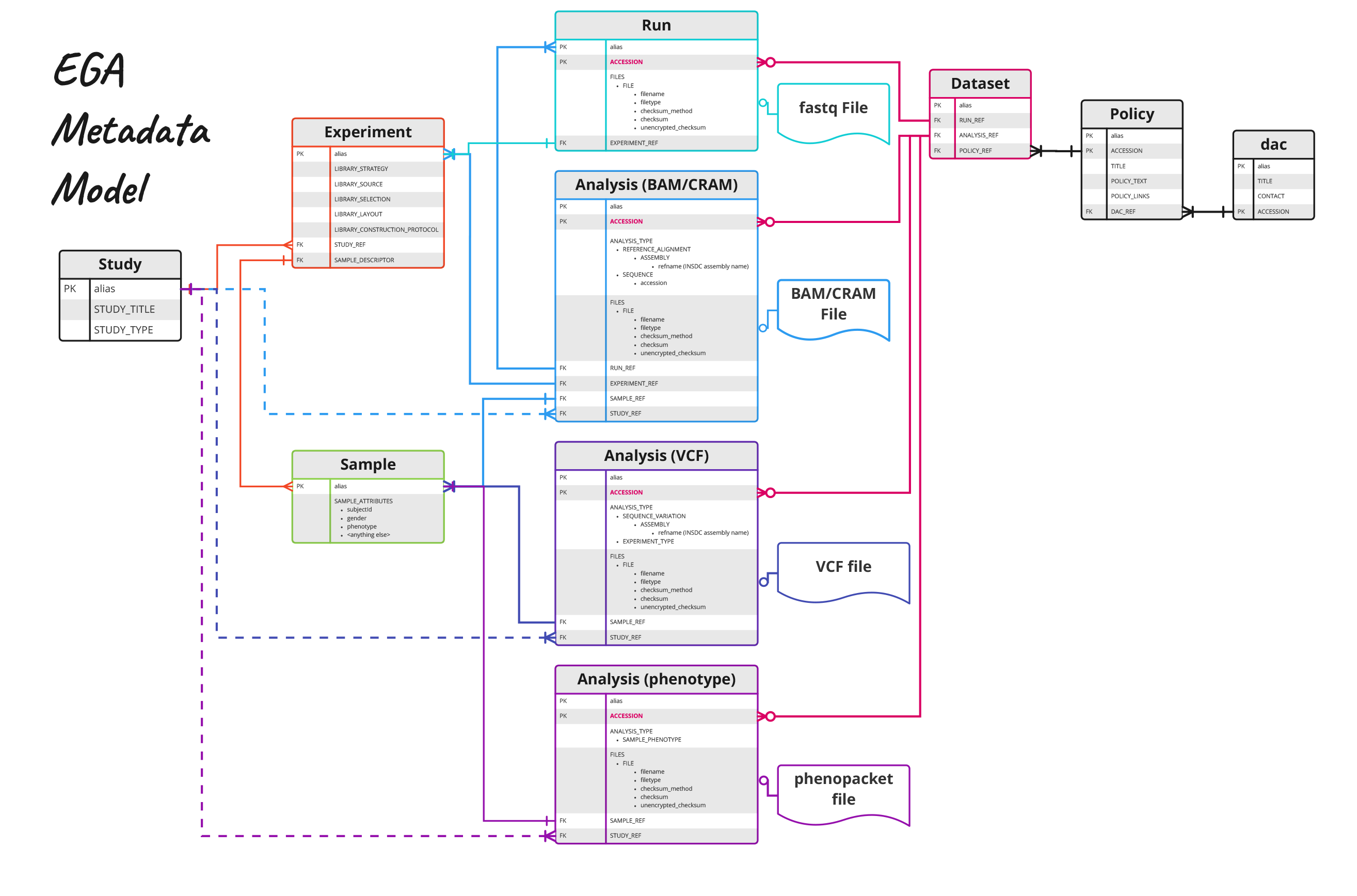
Figure 1. Graphical representation of EGA Metadata Model to represent a sequencing experiment with some recommended fields specified based on EGA example xmls. For comprehensive detail of available fields, see the XML schemas.
Resources:
-
Set of example XMLs created by EGA: https://ega-archive.org/submission/sequence/programmatic_submissions/prepare_xml
-
A somewhat tongue-in-cheek but potentially useful 4 part run through of one user’s experience with how to submit to EGA, mostly programmatically:
- Example of automated data flow to EGA from QIMRB. This transforms their internally stored metadata into the XMLs required for an EGA submission: https://github.com/delocalizer/ega_metadata
EGA Data Files
All files that will be referenced from the Run or Analysis objects must be encrypted before they are transferred to the EGA servers. Encryption is done by using the EGAcryptor software and you need the public key provided by EGA. The files must be checksummed both before and after encryption, and the checksums as well as the method used must be incorporated into the metadata files. File can be transferred using Aspera (faster) or FTP. They don’t generally allow for more than 10TB of data to be uploaded to a submission box at a time, but this can be negotiated with the helpdesk if a higher limit is needed.
Resources:
EGA file upload documentation: https://ega-archive.org/submission/tools/ftp-aspera
Automated encryption and submission of data files from an Amazon S3 bucket to EGA submission box using Aspera, created by UMCCR: https://github.com/umccr/ega-submit
Walk through process: https://github.com/QingliGuo/EGA_Data_submission
Specific software for ICGC members but may be useful/adaptable for other submitters: https://github.com/icgc-dcc/egasub (not actively maintained)
EGA Access Process
Once a dataset of interest is found, a user must apply directly to the data access committee that controls the dataset. There may be different requirements around what information is required to be submitted to apply for access. Any dataset use must comply with the data access policy that is attached to the dataset.
Data access committees will follow their own processes to assess the application, and if approved, contact the EGA to inform them of their decision. Access is then given to a particular EGA account to download the data from their system. The EGA provides a command line program to facilitate downloading of files from the EGA - EGA Download Client.
Download files of interest, can be a subset, particular files/runs/samples? Demonstrate how to use XMLs to subset data and download files/samples of interest.
https://ega-archive.org/download/downloader-quickguide-APIv3
https://github.com/EGA-archive/ega-download-client
Unencrypt files using provided key
https://ega-archive.org/access/data-access
Resources:
EGA data access guide: https://ega-archive.org/access/data-access
Federated EGA
There are currently federated EGA nodes being set up in other countries in Europe such as Finland, Sweden, Germany and Spain. The main driver for this has been to allow for data to remain within the country it is generated.
As part of the Human Genomes Platform Project, leading Australian institutes are looking into the feasibility of setting up a federated node in Australia.
References
- Freeberg, M. A., Fromont, L. A., D’Altri, T., Romero, A. F., Ciges, J. I., Jene, A., Kerry, G., Moldes, M., Ariosa, R., Bahena, S., Barrowdale, D., Barbero, M. C., Fernandez-Orth, D., Garcia-Linares, C., Garcia-Rios, E., Haziza, F., Juhasz, B., Llobet, O. M., Milla, G., … Rambla, J. (2022). The European Genome-phenome Archive in 2021. Nucleic Acids Research, 50(D1), D980–D987. https://doi.org/10.1093/nar/gkab1059
- EGA. (2022). Prepare XMLs - EGA European Genome-Phenome Archive. https://ega-archive.org/submission/sequence/programmatic_submissions/prepare_xml
- Band, G. (2019). Me vs. the EGA part 4: losing again. https://gavinband.github.io/bioinformatics/data/2019/05/15/Me_versus_the_European_Genome_Phenome_Archive_part_four.html
Relevant tools and resources
Skip tool table| Tool or resource | Description | Related pages | Registry |
|---|---|---|---|
| Experimental Factor Ontology (EFO) | The Experimental Factor Ontology (EFO) provides a systematic description of many experimental variables. The scope of EFO is to support the annotation, analysis and visualization of data handled by many groups at the EBI. | Standards/Databases Publication | |
| Phenopackets | GA4GH standard for sharing disease and phenotype information that characterizes an individual person, linking that individual to detailed phenotypic descriptions, genetic information, diagnoses, and treatments. | GA4GH | Publication |
| The European Genome-phenome Archive (EGA) | EGA is a service for permanent archiving and sharing of all types of personally identifiable genetic and phenotypic data resulting from biomedical research projects | BioSample Omics Discovery Index Repositories | Tool info Standards/Databases Training Publication |



