htsget is a data transfer protocol designed to facilitate efficient and secure access to genomic data stored in various formats such as BAM, CRAM, and VCF. The fundamental goal of htsget is to introduce a standardized interface for requesting and delivering genomic data that is not bound by file semantics.
Overview of htsget
htsget (HTTP Sequence GET) is a protocol developed under the Global Alliance for Genomics and Health (GA4GH). It allows clients to request genomic data over HTTP in a manner that is efficient and secure. The protocol is designed to support the retrieval of specific data regions without the need to download entire files, which is crucial for working with large genomic datasets.
What it is not
The protocol does not attempt to provide an end-to-end solution for managing genomic data. Issues around the organization of metadata and data discovery are outside the scope of this protocol. The protocol explicitly does not provide a way to discover the identifiers for valid ReadGroupSets — clients obtain these via some out of band mechanism.
Key Features of htsget
-
Region-Specific Data Retrieval: htsget allows clients to specify genomic regions of interest. This means users can efficiently retrieve only the parts of the data they need, rather than downloading entire files, significantly reducing data transfer times and bandwidth usage.
-
Standardized API: The protocol defines a standard API for requesting data. This standardization ensures interoperability between different systems and tools within the genomics community.
-
Security: The protocol supports secure data transfer mechanisms, ensuring that sensitive genomic data is protected during transmission.
How htsget Works
The key mechanic of the protocol is that the client provides a URL (determined via another discovery service), a preferred format and an optional genomic range via a HTTP(s) GET request (Fig. 1). The server returns a small JSON block with a list of URLs. The client downloads the data from the URLs, concatenates the downloaded data in the order provided by the server to produce the full result of their query.
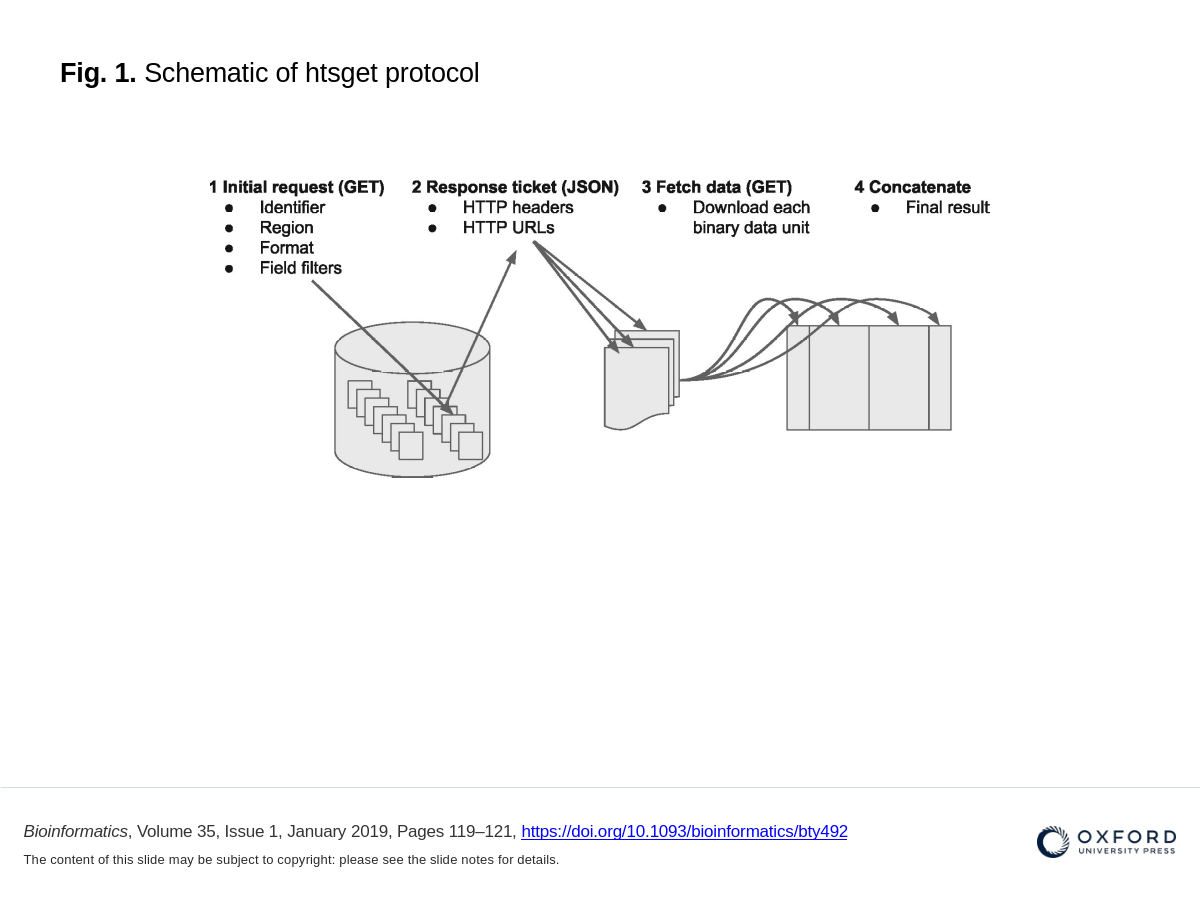
Figure 1. Schematic of htsget protocol
-
Client Request: A client (e.g., a researcher or an analysis tool) makes a request to an htsget server, specifying the genomic regions and data formats they need.
-
Server Response: The htsget server processes the request and generates URLs for the requested data segments. These URLs are provided to the client, often along with metadata about the data segments.
-
Data Retrieval: The client uses the provided URLs to download the specific data segments. This process can be repeated as needed to retrieve additional regions or data formats.
Example Scenario
Suppose a researcher is interested in a specific gene on chromosome 12. Instead of downloading the entire BAM file containing sequencing data for a whole genome, the researcher can use htsget to request only the data for the specific region of chromosome 12. The htsget server will respond with URLs to the specific data segments, which the researcher can then download and analyze.
