Project Description
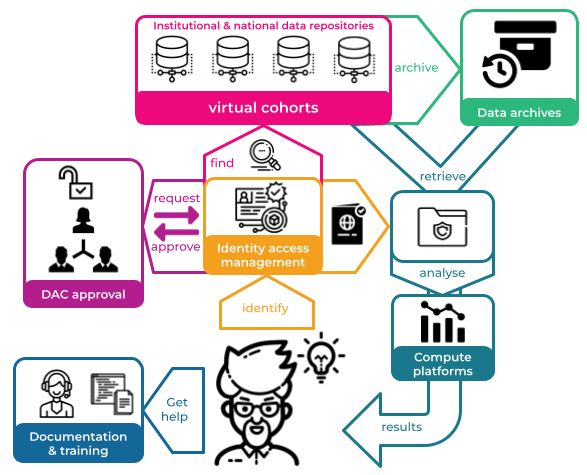
Figure 1. A Researcher’s journey when discovering, accessing and analysing human genomics data.
A researcher looking for Human Genomics research data to analyse needs to go through a complex multi-step process in order to gain access to the data they need. First they need to find data that is of interest to their research, meeting certain criteria for phenotype, demographics or clinical information. Currently, there is no unified way to do this so researchers need to search through multiple data and publication repositories and try to bring together data from many different sources.
Due to the sensitive nature of human genomics data, researchers need to prove their identity as a bona fide researcher in order access detailed information in repositories. Currently, this is largely isolated to each different repository, meaning users need to register independently and keep track of their various identities. It also makes it difficult for the owners of the service to verify the identity of each user as bona fide researchers.
Once they find data of interest, they often need to request access through a data access committee (DAC). This is often an ad-hoc manual process that involves back and forth emails between the data requester, data custodians, data administrators and data access committee members. The ad-hoc manual nature makes it difficult to track, audit and report on at a later date.
After their request is approved through the DAC, a research will often then need to retrieve the data from a repository or archive. In Australia, there are no national archives, so this often means downloading large data files from North America or Europe which can be time-consuming and prone to drop-outs.
The HGPP seeeks to improve this researcher journey by discovering and testing the best global technologies and laying the foundation to implement them at a national scale in the five sub-project areas:
- Virtual Cohorts - Discovering and assembling cohorts of interest across multiple repositories
- Data Access Committee Support - Implementing DAC management software to streamline data access requests and approvals
- Federated Identity & Access Management - Establishing trusted identities linked to institutional accounts that can be used for national genomic services and resources
- Data & Metadata Archiving - Investigating the feasibility of establishing a national human genomics archive in Australia
- Communications, Documentation & Training - Ensuring all implemented solutions are communicated and there is support to use them
Knowledge discovery
Requirements gathering User Needs Identifying Gaps and pilot solutions Deciding on pilot to implement and assess
Pilot implementations
After knowledge gathering we proceeded with testing out pilot solutions for each sub-project as shown below.
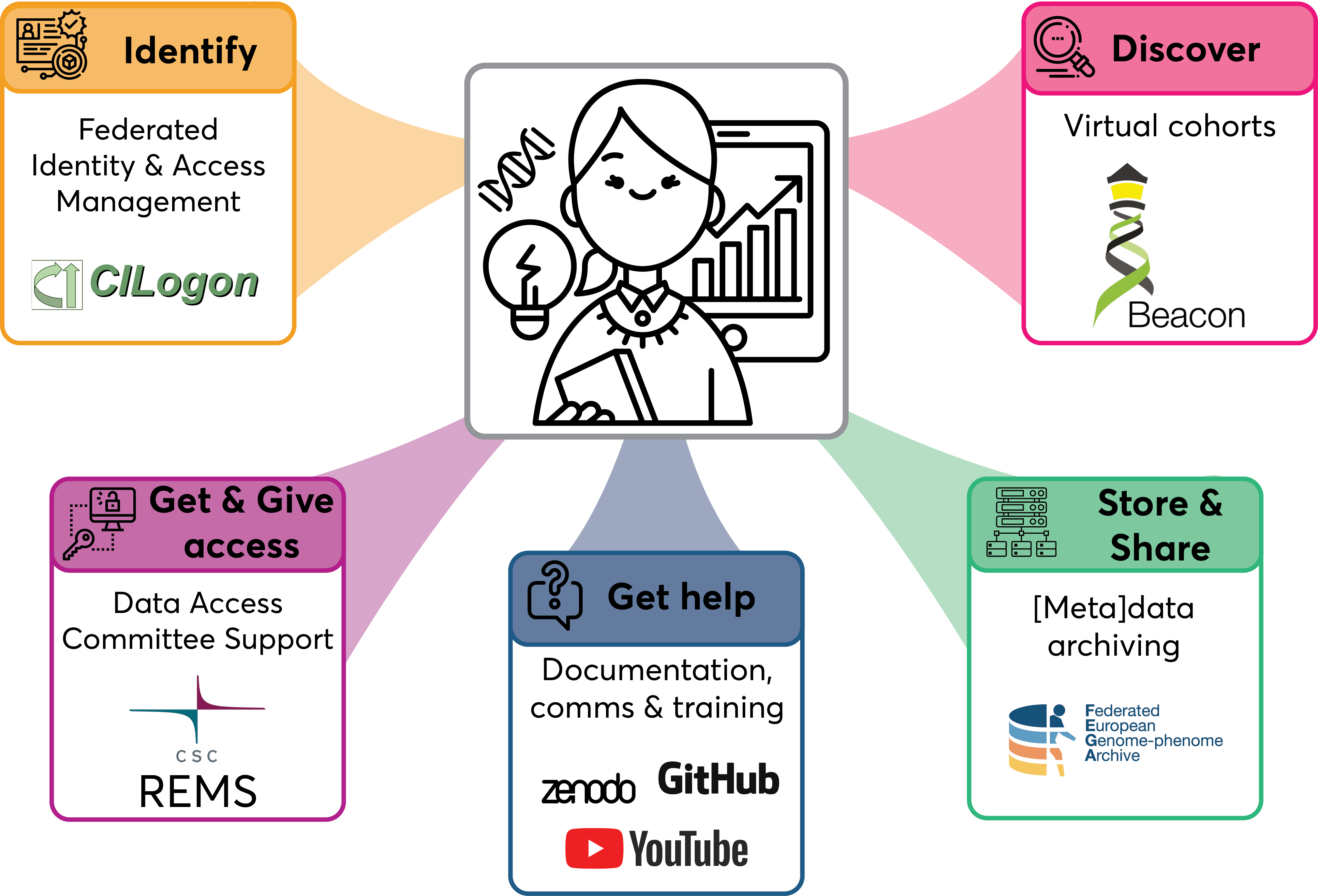
Figure 2. Pilot solutions tested in the HGPP.
The findings from this pilot phase are summarised in the pilot implementation reports for each sub-project
Project outputs
Pilot Discovery reports
Pilot Implementation reports
(Coming Soon…)
Conference talks and posters
-
Project overview poster presented at ELIXIR All Hands 2022 (Shadbolt et al., 2022)
-
Project overview poster presented at eResearch Australasia 2022 (Shadbolt et al., 2022)
-
Beacon network poster presented at ABACBS 2022 (Taouk et al., 2022)
-
Beacon network UI poster presented at ABACBS 2022 (So et al., 2022)
-
Beacon network UI slides from oral presentation given at COMBINE 2022 (So et al., 2022)
-
Project overview as part of poster presented at the International Congress of Genetics and Genomics (Shadbolt et al., 2023)
Webinars
-
Protection of genomic data and the Australian Privacy Act: is genomic data ‘personal information’? (Australian BioCommons, 2022)
-
Genomic data - improving discovery and access management (Australian BioCommons, 2023)
Flyer
Acknowledgements
The HGPP received investment from the NCRIS-enabled ARDC infrastructure under investment identifier https://doi.org/10.47486/PL032 as well as being funded through BioPlatforms Australia. Contributions are also made from each partner organisation: QIMR Berghofer Medical Research Institute, The University of Melbourne Centre for Cancer Research, Garvan Institute for Medical Research, ZERO Childhood Cancer & the Children’s Cancer Institute, Australian Genomics, Melbourne Genomics Health Alliance, National Computational Infrastructure and the Australian Access Federation.
Icons in Figures 1 and 2 from the Noun Project: Computing, search, cloud by Flatart, database by Start Up Graphic Design, Analyse by Taylan Sentürk, computing by Phonlaphat Thongsriphong, identified by Tippawan Sookruay, group by Gregor Cresnar, group by Oksana Latysheva, Data File by Blangcon, Unlock by Arthur Shlain, archive by Adrien Coquet, support by Komkrit Noenpoempisut, documentation by lastspark, Scientist by Maxim Kulikov, Immigration Approval by Ary Prasetyo, identified by Tippawan Sookruay, Data access by monkik, Help by Gregor Cresnar, DNA by LAFS, Scientist by Amethyst Studio, Data Sharing by Vectors Point.
References
- Shadbolt, M., Holliday, J., Winter, U., Manos, S., Christiansen, J., Lonie, A., & Pope, B. (2023). Advancing Human Genomics Data Sharing In Australia: Highlights From The Australian BioCommons. https://doi.org/10.5281/zenodo.8137358
- Australian BioCommons. (2023). Genomic data - improving discovery and access management. https://www.youtube.com/watch?v=9SD6gpjDGWE
- Taouk, K., Lin, A., Wong-Erasmus, M., Cowley, M., Boughtwood, T., Christiansen, J., Copty, J., Ravishankar, S., Davies, K., Downton, M., Druken, K., Evans, B., Gaff, C., Gilbert, A., Hall, C., Hobbs, M., Hofmann, O., Holliday, J., Kaplan, W., … Syed, M. (2022). Establishing a national Beacon version 2 network for real-time genomics data discovery. https://doi.org/10.5281/zenodo.7402705
- So, D., Nguyen, R., Do, J., Kamarinos, Z., Lin, A., Cowley, M., Syed, M., Taouk, K., & Wong-Erasmus, M. (2022). Deploying a User Interface for Sharing Federated Genomic and Phenotypic Data using the Beacon v2 Protocol. https://doi.org/10.5281/zenodo.7416545
- So, D., Nguyen, R., Do, J., & Kamarinos, Z. (2022). Developing a User Interface for Sharing Federated Genomic and Phenotypic Data Using the Beacon v2 protocol. https://doi.org/10.5281/zenodo.7416582
- Cowley, M., Downton, M., Holliday, J., Kummerfeld, S., Leonard, C., Lin, A., Pope, B., San Kho Lin, V., Ravishankar, S., Shadbolt, M., Syed, M., Taouk, K., & Wong-Erasmus, M. (2022). Virtual Cohort Assembly Discovery Phase Report: National Community Needs & Candidate Solutions. Zenodo. https://doi.org/10.5281/zenodo.7439886
- Shadbolt, M., Boughtwood, T., Christiansen, J., Copty, J., Cowley, M., Davies, K., Downton, M., Druken, K., Evans, B., Gaff, C., Gilbert, A., Hall, C., Hobbs, M., Hofmann, O., Holliday, J., Kaplan, W., Koufariotis, R., Kummerfeld, S., Leonard, C., … Wood, S. (2022). Enhancing Australia’s capability for secure and responsible sharing of human genomics research data. https://doi.org/10.5281/zenodo.7242979
- Carnuccio, P., Cowley, M., Davies, K., Downton, M., Dumevska, B., Holliday, J., Kummerfeld, S., Lin, A., Monro, D., Patterson, A., Pope, B., Ravishankar, S., Robinson, A., Scullen, J., Shadbolt, M., Syed, M., Wood, S., & Wong-Erasmus, M. (2022). Human Genomes Platform Project: Federated Identity and Access Management (IAM) Discovery Phase Report. Zenodo. https://doi.org/10.5281/zenodo.6644009
- Carnuccio, P., Cowley, M., Davies, K., Druken, K., Holliday, J., Kummerfeld, S., Monro, D., Patterson, A., Pearson, J., Pope, B., Scullen, J., Shadbolt, M., Wong-Erasmus, M., & Wood, S. (2022). Human Genomes Platform Project: DAC Automation Discovery Phase Report. Zenodo. https://doi.org/10.5281/zenodo.6644050
- Shadbolt, M., Boughtwood, T., Christiansen, J., Copty, J., Cowley, M., Davies, K., Downton, M., Druken, K., Evans, B., Gaff, C., Gilbert, A., Hall, C., Hofmann, O., Holliday, J., Kaplan, W., Koufariotis, R., Kummerfeld, S., Leonard, C., Lin, A., … Wood, S. (2022). National and international collaboration to facilitate human genomics data sharing in Australia: The Human Genomes Platform Project. F1000Research, 11. https://doi.org/10.7490/f1000research.1118989.1
- Australian BioCommons. (2022). Protection of genomic data and the Australian Privacy Act: is genomic data ‘personal information’? https://www.youtube.com/watch?v=Iaei-9Gu-AI



