Structure-based function annotation
Overview
Teaching: 5 min
Exercises: minQuestions
Objectives
Learn how to obtain a functional understanding of an uncharacterised protein using its structure.
Genome annotation and why structure matters
Despite the explosion in genomic data, a large fraction of genes remain annotated only as hypothetical proteins. Traditional genome annotation relies on sequence similarity, but this approach breaks down when homologous sequences are missing or highly diverged.
In this session, we’ll walk through a case study from the genome of Candidatus Protochlamydia naegleriophila.
-
Before commencing the exercise, navigate to the relevant working directory:
cd $MYSCRATCH/2025-ABACBS-workshop/exercises/exercise1/ -
Download the Genbank genome annotation file:
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/499/655/GCF_001499655.1_PNK1/GCF_001499655.1_PNK1_genomic.gbff.gz -
Count the number of
hypothetical proteinannotations:zgrep "hypothetical" GCF_001499655.1_PNK1_genomic.gbff.gz | wc -l -
Count the number of
protein codinggenes:zgrep "protein_id" GCF_001499655.1_PNK1_genomic.gbff.gz | wc -l
Review
Note that
802/2516(>30%) of the protein-coding genes do not have functional annotations.The good news: protein structures are often more conserved than sequences, and they are tightly linked to function. Recent advances in structure prediction (using tools we’ll explore today) allow us to analyse structures at genomic scale. By comparing predicted 3D structures to known proteins, we can uncover functional relationships that sequence-based approaches miss.
Representative example: uncovering the function of a “hypothetical protein”
We’ll examine a gene annotated as a conserved hypothetical protein (locus tag PNK_0205) and explore how protein structure-based annotation can yield new functional hypotheses.
This case study is taken from:
- Litfin,T. et al. (2025) Ultra-fast and highly sensitive protein structure alignment with segment-level representations and block-sparse optimization.
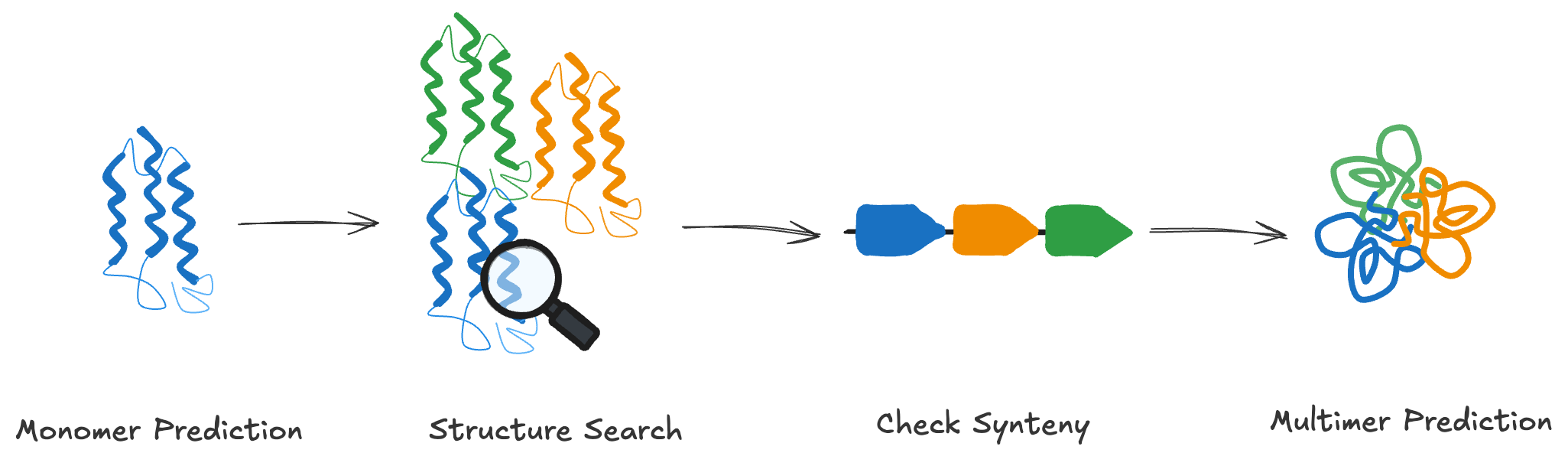
Strategy
- Predict the 3D structure of the target protein as a monomer.
- Search for similar annotated structures.
- Compare gene neighborhoods of our protein and the matched structure.
- Predict the complex structure of potential interaction partners based on known functional associations.
Key Points
Many genes do not have functional annotations.
Predicted structures can provide important clues about function.