Leveraging peak Australian compute to enable workflows for predictive structural biology at scale -- 2025 ABACBS workshop: Setup
Key requirements
The main requirement for this workshop is a personal computer with a web browser and Visual Studio Code (VSCode). This equipment will allow you to follow the online materials and login to a facility with the required software stack.
You should also be familiar with bash scripting. This is an introduction to Bash that you might find useful: The Unix Shell: Summary and Setup
Suggested
Recommended for use after the workshop
- Molecular Viewer installed (PyMOL/ChimeraX)
Set up your computer
In this workshop, we will use the Setonix HPC at Pawsey Supercomputing Research Centre (Perth, WA).
Each participant will be provided with their training account and password at the workshop.
Before the workshop, you must have the following:
VSCodeinstalled.- The
Remote - SSHVSCode extension installed.
Below, you will find instructions on how to set up VSCode.
Installing Visual Studio Code
Visual Studio Code (VSCode) is a versatile code editor that we will use for the
workshop. We will use VSCode to connect to the HPC, download and execute the Proteinfold workflow, monitor jobs on the HPC, and edit, view and download files.
-
Download
VSCodeby following the installation instructions for your local Operating System. -
Open
VSCodeto confirm it was installed correctly.
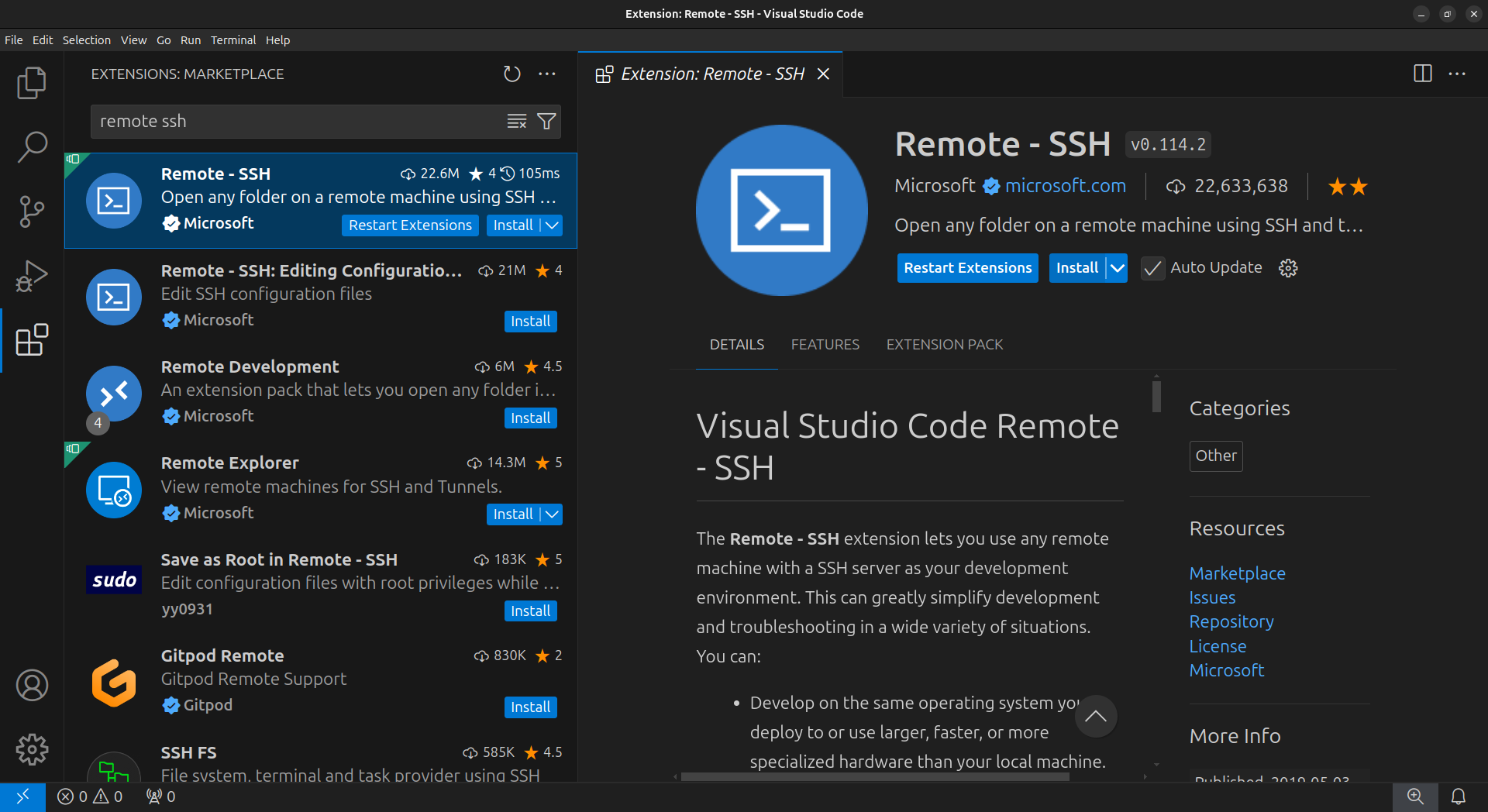
Installing the Remote - SSH VScode extension
-
In the VSCode sidebar on the left, click on the extensions button (four blocks)
-
In the Extensions Marketplace search bar, search for
remote ssh. Select “Remote - SSH”
You have now configured VSCode for the workshop!
Acknowledgements
Setup content copied and adapted with permission from a Sydney Informatics Hub (SIH, University of Sydney) Nextflow and HPC workshop.