Using structures to annotate function
Overview
Teaching: 5 min
Exercises: 5 minQuestions
Objectives
Learn about services (Foldseek and SPfast) for searching large structure databases.
Interpret functional consequences of structural similarity.
Find supporting evidence for functional annotations.
Database search with predicted structures
Now that we have our high-confidence predicted structure, we can use it to search a large database of annotated structures.
Similar annotated structures could provide a hypothesis about the likely function of our target protein.
Download structure predictions
-
Download the
sample0_alphafold2.pdbfile located in theoutput/alphafold2/standard/sample0/directory. You can find the file by navigating to theoutput/alphafold2/standard/sample0/directory in the VS-code file browser on the left-hand panel. -
Right-click the
sample0_alphafold2.pdbfile and selectDownload.
Foldseek
The Foldseek server is extremely fast and useful for identifying structural matches across a range of experimental and predicted structure databases.
Careful
When deciding to send data to an external service, it is important to understand:
- How the data will be handled (e.g. transferred, processed, stored, secured), and
- If it is appropriate to submit the data to the service you are intending to use.
Do not send sensitive data to an external service without understanding potential risks.
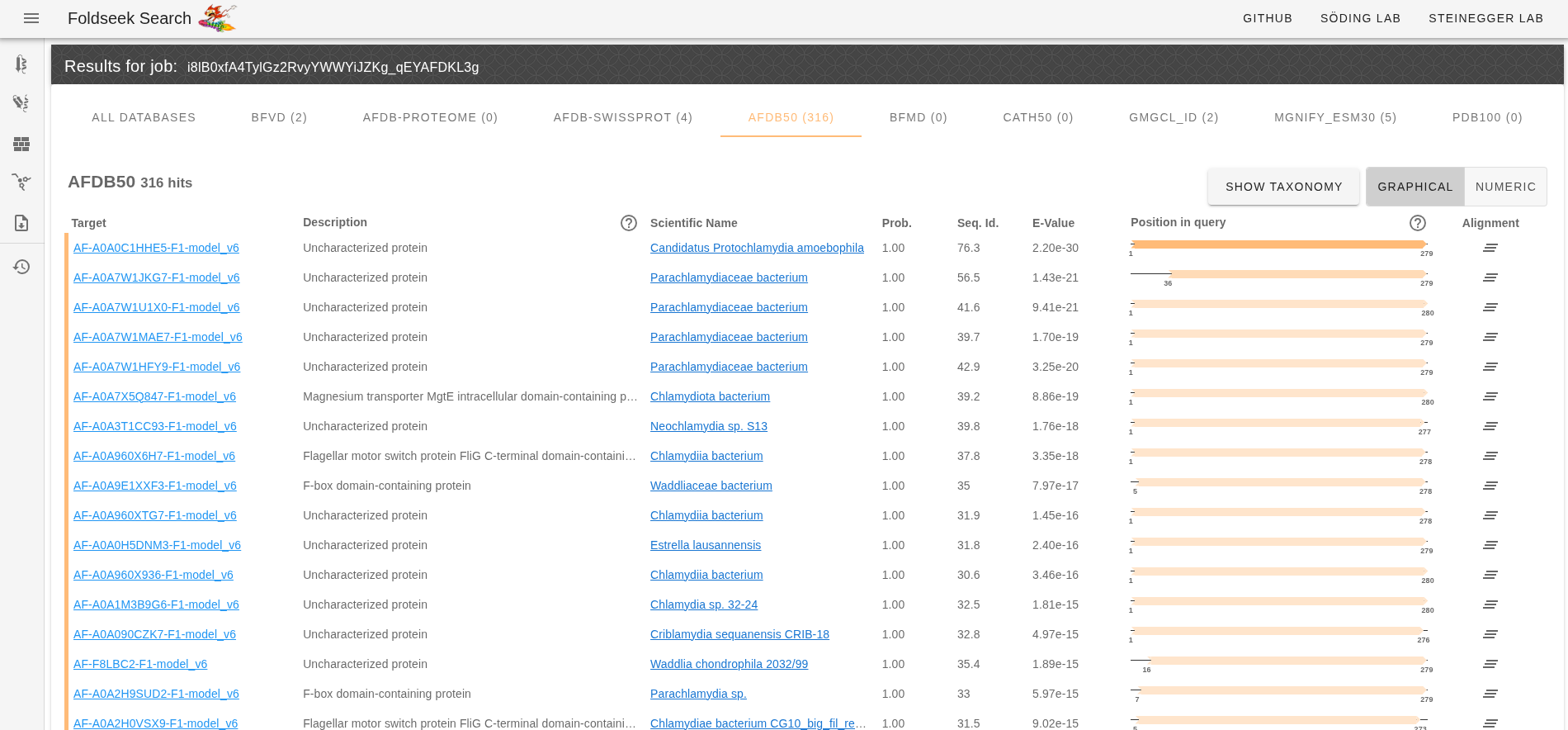
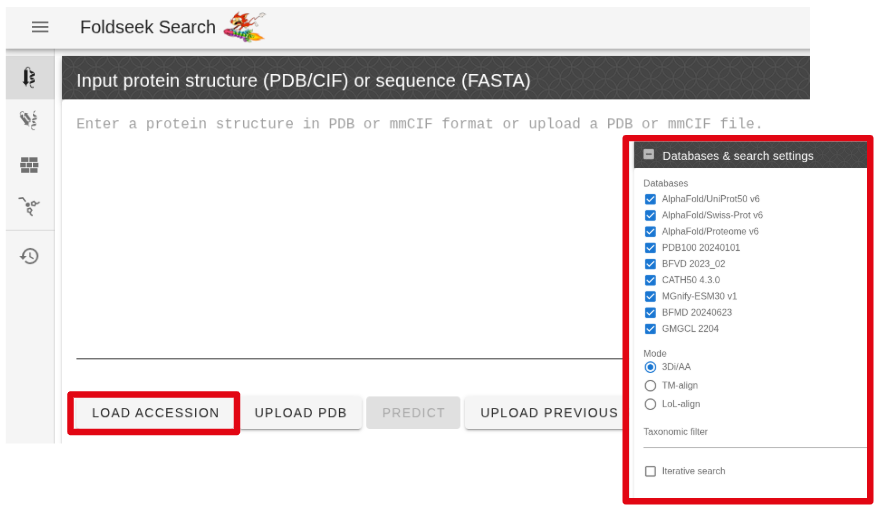
- Upload our predicted PDB structure to the Foldseek server and search for similar structures. Ensure that the check-boxes for all databases are selected (see screenshot below).
Input form
Results
- Browse the tabs to see the top hits in various protein structure databases.
- The most similar proteins in AFDB50 are also uncharacterised proteins.
- There are some hits to proteins with various annotations (MgtE, FliG, F-box).
- Many fringe hits are annotated with FliG, YscK and SctK - a component of the Type III secretion system.
Careful
- Structural similarity does NOT guarantee a related function.
- Shared structural scaffolds can sometimes adopt highly divergent functions.
- We can look for complementary evidence to support structure-based annotations.
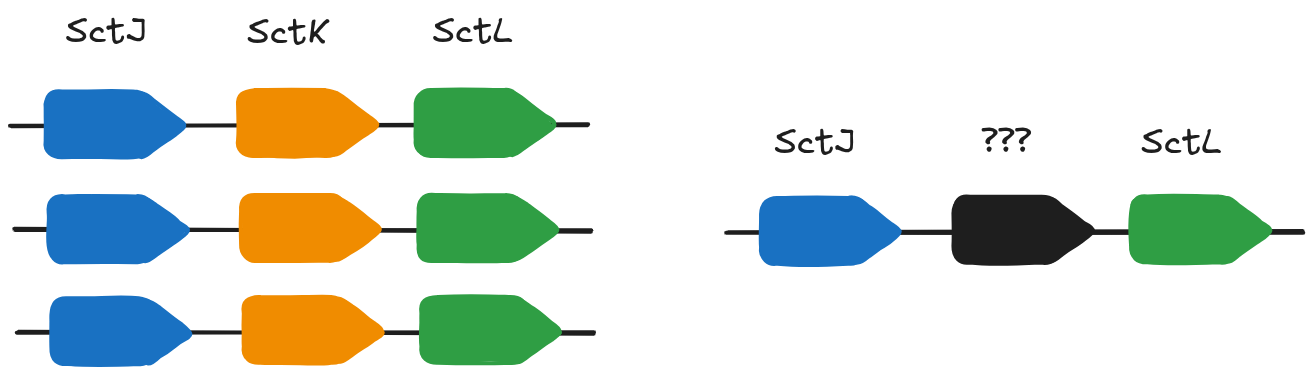
Synteny
T3SS genes are often found in operons. In genomes where it is annotated, SctK is overwhelmingly found between SctL and SctJ.
-
Return to our original assembly and look at the gene neighbors of our target locus (PNK_0205).
Gene Neighborhood
gene complement(256361..257014) /gene="sctL" /locus_tag="PNK_0204" CDS complement(256361..257014) /gene="sctL" /locus_tag="PNK_0204" /function="Flagellar biosynthesis/type III secretory pathway protein" /codon_start=1 /transl_table=11 /product="putative type III secretion protein SctL" /protein_id="CUI15842.1" /translation="MSKKFFSLIYGDQIHTAPETKVIPADSFSVLQDASQVLELIKQD AEKYRMQVVKESEQLKEHAEKEGYEEGFKKWAEHLVNLEKEIEKVHQELQQLVIPVAL KAAKKIVGKEIELSEDVIVDIVASNLKAVAQHKKVTIFVNKKDLDVLDKNKPRLRDLF ESLESLSIRPRDDVASGGCIIETEIGIINAQLEHRWRVLEKAFEGLVKTSPEPEKGS" gene complement(257017..257859) /locus_tag="PNK_0205" CDS complement(257017..257859) /locus_tag="PNK_0205" /codon_start=1 /transl_table=11 /product="conserved hypothetical protein" /protein_id="CUI15843.1" /translation="MDKRGWMMLRVFINCYNPKAGEALLKFLPQEEVQAVLSQDIRST DLTPILYQPQKLLERMHYSWIEPLLGGFPEKLHPLVMAALTQEQISGLNPVIAPSTLS NPVKTFIINQLYTLLKADEHLPYDYLPETDLSPLGTWSKARLTELIDFLGLHDLASEM RHIVDKNQLKNIYTSLSSKQFYYLKVCLHQKEILSVPKLGIDPSKRDSTKLKRIVHRR GLLRLGKALCGQHPDFVWYLAHTLDTGRGKLILNAYQPESVPQVTSFLKGQVLNLMNF LKSE" gene complement(257888..258895) /gene="sctJ" /locus_tag="PNK_0206" CDS complement(257888..258895) /gene="sctJ" /locus_tag="PNK_0206" /function="Type III secretory pathway, lipoprotein EscJ" /codon_start=1 /transl_table=11 /product="type III secretion lipoprotein SctJ" /protein_id="CUI15844.1" /translation="MKINCVAARTSIYRFLHQLMVFITLVSVLTSCESRRVIVNGLEE KEANEILVFLSTKGINATKVQAATEGGGGGKGILWNISVEETQANEAMALLNQVGLPR RRGQNLLGIFANTSLVPSGMQEKIRYQAGLAEQIASTIRKIDGVLDADVQISFPDEDP LNPNAPKQKITASVYVKHNGVLDDPNAHLTTRIKRLVSGSVNGLDYDNVTVIGDKARY GETPLGGLGGSLGDEEKQYVNVWSIVLAKDSLSRFRIIFFAFTISLVLLLLALIWLLW KFLPLLKKVGGFKQLLSFHPIQLGDIATEAKAPEATDAKKEEKAKKSEDDTANQGIDE T"
PNK_0205 is between the SctL and SctJ genes which are also components of the T3SS machinery.
The gene neighborhood of our uncharacterised gene is consistent with our protein structure based annotation.
Note:
Structure-based annotation is on the roadmap for another nf-core pipeline - proteinannotator
Key Points
Similar structures can often share a similar function.
Structure-based annotation should be supported by complementary evidence.